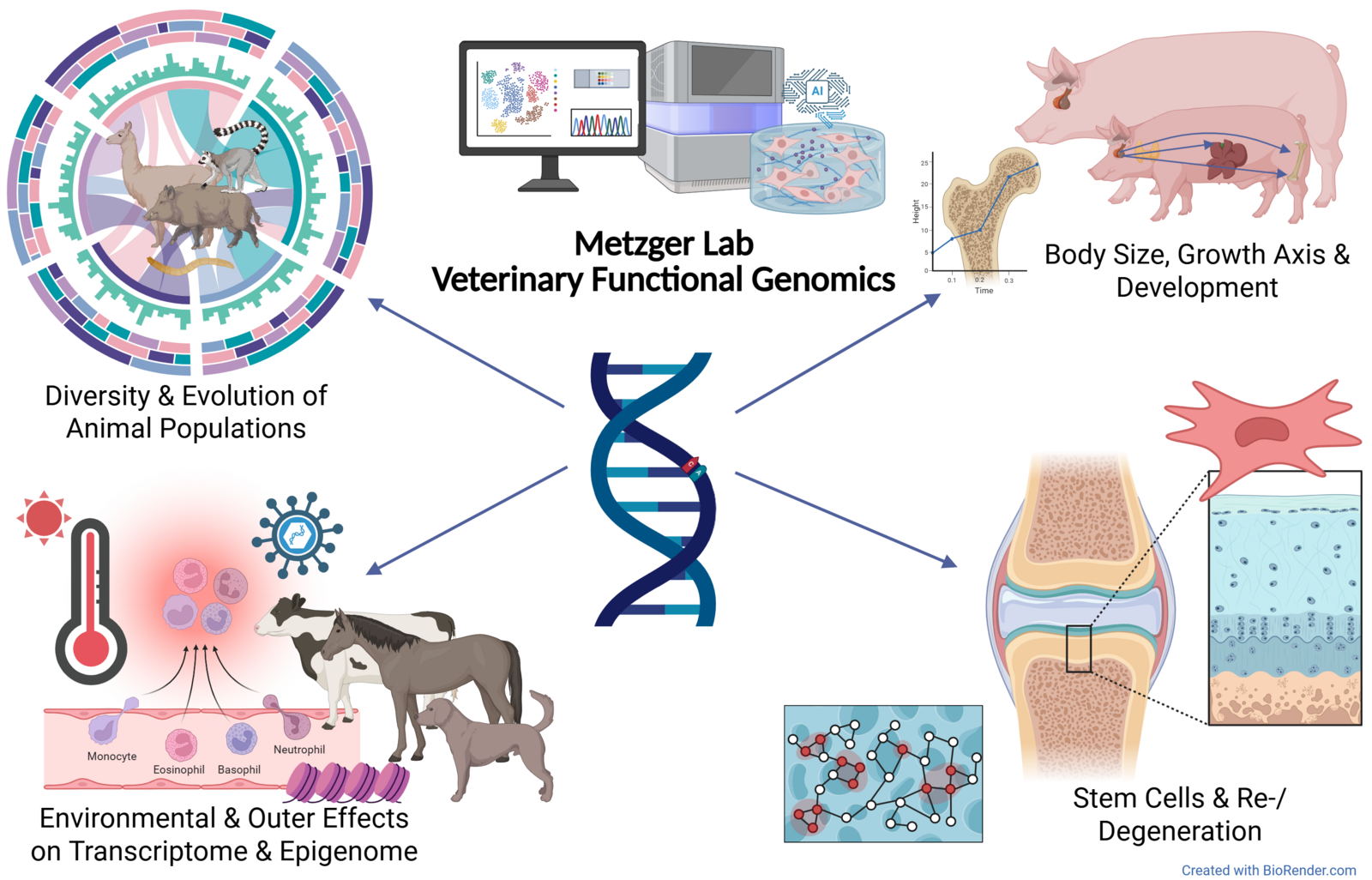

Our research
Our research group investigates genomic variation and architecture as well as the function and regulation of genomic elements in veterinary medicine. We use advanced OMICs technologies, including RNA-seq, ATAC-seq, and Hi-C, alongside DNA language models to decode regulatory patterns and better understand complex genotype-phenotype relationships. A central focus lies on growth and skeletal development processes, particularly at the growth plate, in domestic animals such as pigs. We also explore the role of stem cells in tissue regeneration and degeneration, and examine functional and epigenetic mechanisms underlying diseases influenced by environmental factors like infections or temperature stress. Another key area of our work is the analysis of evolutionary processes in domestic species, with a particular focus on selection signatures and the genomic consequences of artificial selection. Our research integrates in vitro models with single-cell analyses and bioinformatics to uncover functional mechanisms at the molecular level.
Head of working group:
Research projects
Latest research projects (Selection; DFG projects: see DFG GEPRIS) in functional genomics
Growth & Disease
- MEASURE: Multi-omics studies for body size in the animal model – the genetic architecture of pig body size (DFG-Heisenberg-Studie)
- 3D-DRIVE: Dynamic 3D genome reorganization of porcine growth
- SKIP: Functional analysis of skeleton-modifying genes and interactive gene expression networks
- SPAID-research: Functional studies for Shar-Pei Autoinflammatory Disease (see here)

Genomstruktur & Evolution
- WORMICs: 3D Genomics of the mealworm
-
Structural variation related to domestication and selection of livestock breeds
A full list of thrid-party funded projects can be found here: TiHo-Datenbank für Forschungsprojekte
Publications
Selected Publications
Genome & Selection Signatures:
- Berghöfer J, Khaveh N, Mundlos S, Metzger J (2022) Simultaneous testing of rule- and model-based approaches for runs of homozygosity detection opens up a window into genomic footprints of selection in pigs. BMC Genomics 23 (1): 1-26. https://doi.org/10.1186/s12864-022-08801-4 (open access)
Growth Deficiency & Miniaturization:
- Halecker, S., Metzger, J., Strube, C., Krabben, L., Kaufer, B. & Denner, J. (2021) Virological and Parasitological Characterization of Mini-LEWE Minipigs Using Improved Screening Methods and an Overview of Data on Various Minipig Breeds. Microorganisms 9, 9122617. https://doi.org/10.3390/microorganisms9122617
- Schachler K, Minx J-O, Sürie C, Distl O, Metzger J. (2020) Genetic characterisation of Mini-LEWE as resource population for experimental research. Berliner und Münchener Tierärztliche Wochenschrift 133, DOI: 10.2376/1439-0299-2020-15
- Metzger J, Rau J, Naccache F, Bas Conn L, Lindgren G, Distl O. (2018) Genome data uncover four synergistic key regulators for extremely small body size in horses. BMC Genomics 19(1):492. https://doi.org/10.1186/s12864-018-4877-5 (open access)
- Metzger J, Gast AC, Schrimpf R, Rau J, Eikelberg D, Beineke A, Hellige M, Distl O (2017) Whole-genome sequencing reveals a potential causal mutation for dwarfism in the Miniature Shetland pony. Mammalian Genome 28, 143-151. https://doi.org/10.1007/s00335-016-9673-4
- Metzger J, Distl O (2020) Book section: Genetics of Equine Orthopedic Disease. Veterinary Clinics of North America: Equine Practice, 36 (2): 289-301, Edited by Carrie J. Finno. https://doi.org/10.1016/j.cveq.2020.03.008
Transcriptome and Multi-OMICs Studies in Pigs & Other Livestock:
- Khaveh N, Schachler K, Berghöfer J, Jung K, Metzger J (2023) Altered hair root gene expression profiles highlight calcium signaling and lipid metabolism pathways to be associated with curly hair initiation and maintenance in Mangalitza pigs. Frontiers in Genetics, 14: 1184015. https://doi.org/10.3389/fgene.2023.1184015 (open access)
- Schachler K, Distl O, Metzger J (2020) Tracing selection signatures in the pig genome gives evidence for selective pressures on a unique curly hair phenotype in Mangalitza. Scientific Reports, 10 (1): 1-12. https://doi.org/10.1038/s41598-020-79037-z (open access)
- Metzger, J, Schrimpf, R, Philipp, U & Distl, O (2013) Expression Levels of LCORL Are Associated with Body Size in Horses. PLoS ONE 8, https://doi.org/10.1371/journal.pone.0056497 (open access)
For additional publications, see Google Scholar
Career opportunities
We are always looking for highly motivated scientists with an interest/expertise in the fields of genetics, genomics, stem cell development, and bioinformatic methods.
If you are interested, please send an application (cover letter, resume, recommendation letters from two mentors) as a PDF to: julia.metzger@tiho-hannover.de


